Computer simulations help us understand COVID-19
Computer simulations help us understand COVID-19
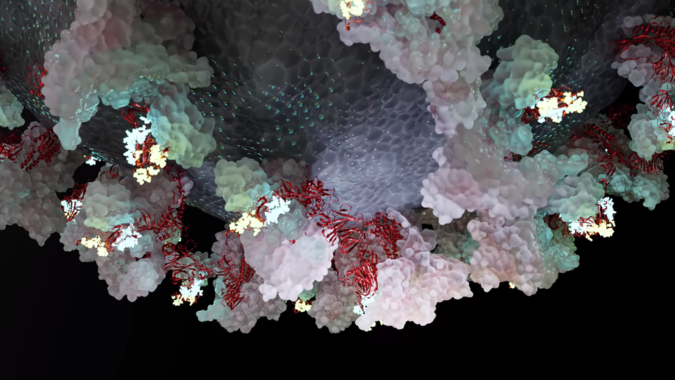
NVIDIA, which is known for its graphics cards, presented an extremely spectacular simulation, which is built upon the achievements of the folding@home initiative. The interactive molecular dynamics presentation shows the transformation of the spike protein present on the surface of COVID-19, as it moves from the “closed” state into the “open” state. This conformational change of the protein is responsible for the binding of the virus to human cells.
In the spring of 2020, KIFU announced that it had joined the folding@home initiative – which intends to map the protein structure of the coronavirus to help with the fight against the pandemic – by offering the computing capacities of the two NVIDIA V1000 graphics processors (GPU) of the SuperDome Flex tool purchased for the project “Developing Earth Monitoring Data Infrastructure and Services” of the Earth Monitoring Information System (FIR). Considering the capacities that were made available, KIFU is only one of the more than 2.7 million contributors but is among the top 25% performers, i.e., those processing the largest part of the computing jobs.
The international initiative folding@home is supported by an open community cooperation program and had considerable achievements during the past years in the field of cancer research, and in understanding the causes of neurological diseases, and also contributed to the development of the antidote to Ebola.

